PSM with MI
The tutorial provides a detailed walkthrough of implementing Propensity Score Matching (PSM) combined with Multiple Imputation (MI) in a statistical analysis, focusing on handling missing data and mitigating bias in observational studies.
The initial chunk is dedicated to loading various R packages that will be utilized throughout the tutorial. These libraries provide functions and tools that facilitate data manipulation, statistical modeling, visualization, and more.
Problem Statement
Logistic regression
Perform multiple imputation to deal with missing values; with 3 imputed datasets, 5 iterations,
fit survey featured logistic regression in all of the 3 imputed datasets, and
obtain the pooled OR (adjusted) and the corresponding 95% confidence intervals.
-
Hints
- Use the covariates (listed below) in the imputation model.
- Imputation model covariates can be different than the original analysis covariates. You are encouraged to use variables in the imputation model that can be predictive of the variables with missing observations. In this example, we use the strata variable as an auxiliary variable in the imputation model, but not the survey weight or PSU variable.
- Also the imputation model specification can be modified. For example, we use pmm method for bmi in the imputation model.
- Remove any subject ID variable from the imputation model, if created in an intermediate step. Indeed ID variables should not be in the imputation model, if they are not predictive of the variables with missing observations.
Predictive Mean Matching:
The “Predictive Mean Matching” (PMM) method in Multiple Imputation (MI) is a widely used technique to handle missing data, particularly well-suited for continuous variables. PMM operates by first creating a predictive model for the variable with missing data, using observed values from other variables in the dataset. For each missing value, PMM identifies a set of observed values with predicted scores that are close to the predicted score for the missing value, derived from the predictive model. Then, instead of imputing a predicted score directly, PMM randomly selects one of the observed values from this set and assigns it as the imputed value. This method retains the original distribution of the imputed variable since it only uses observed values for imputation, and it also tends to preserve relationships between variables. PMM is particularly advantageous when the normality assumption of the imputed variable is questionable, providing a robust and practical approach to managing missing data in various research contexts.
Propensity score matching (Zanutto, 2006)
- Use the propensity score matching as per Zanutto E. L. (2006)’s recommendation in all of the imputed datasets.
- Report the pooled OR estimates (adjusted) and corresponding 95% confidence intervals (adjusted OR).
Data and variables
Analytic data
The analytic dataset is saved as NHANES17.RData.
Variables
We are primarily interested in outcome diabetes and exposure whether born in the US (born).
Variables under consideration:
- survey features
- PSU
- strata
- survey weight
- Covariates
- race
- age
- marriage
- education
- gender
- BMI
- systolic blood pressure
Pre-processing
The data is loaded and variables of interest are identified.
load(file="Data/propensityscore/NHANES17.RData") # read data
ls()
#> [1] "analytic" "analytic.with.miss"
dim(analytic.with.miss)
#> [1] 9254 34
vars <- c("ID", # ID
"psu", "strata", "weight", # Survey features
"race", "age", "married","education","gender","bmi","systolicBP", # Covariates
"born", # Exposure
"diabetes") # OutcomeSubset the dataset
The dataset is then subsetted to retain only the relevant variables, ensuring that subsequent analyses are focused and computationally efficient.
Inspect weights
The weights of the observations are inspected and adjusted to avoid issues in subsequent analyses.
Recode the exposure variable
The exposure variable is recoded for clarity and ease of interpretation in results.
variable types
Variable types are set, ensuring that each variable is treated appropriately in the analyses.
Inspect extent of missing data problem
A visualization is generated to explore the extent and pattern of missing data in the dataset, which informs the strategy for handling them.
require(DataExplorer)
plot_missing(dat.with.miss)
#> Warning: `aes_string()` was deprecated in ggplot2 3.0.0.
#> ℹ Please use tidy evaluation idioms with `aes()`.
#> ℹ See also `vignette("ggplot2-in-packages")` for more information.
#> ℹ The deprecated feature was likely used in the DataExplorer package.
#> Please report the issue at
#> <https://github.com/boxuancui/DataExplorer/issues>.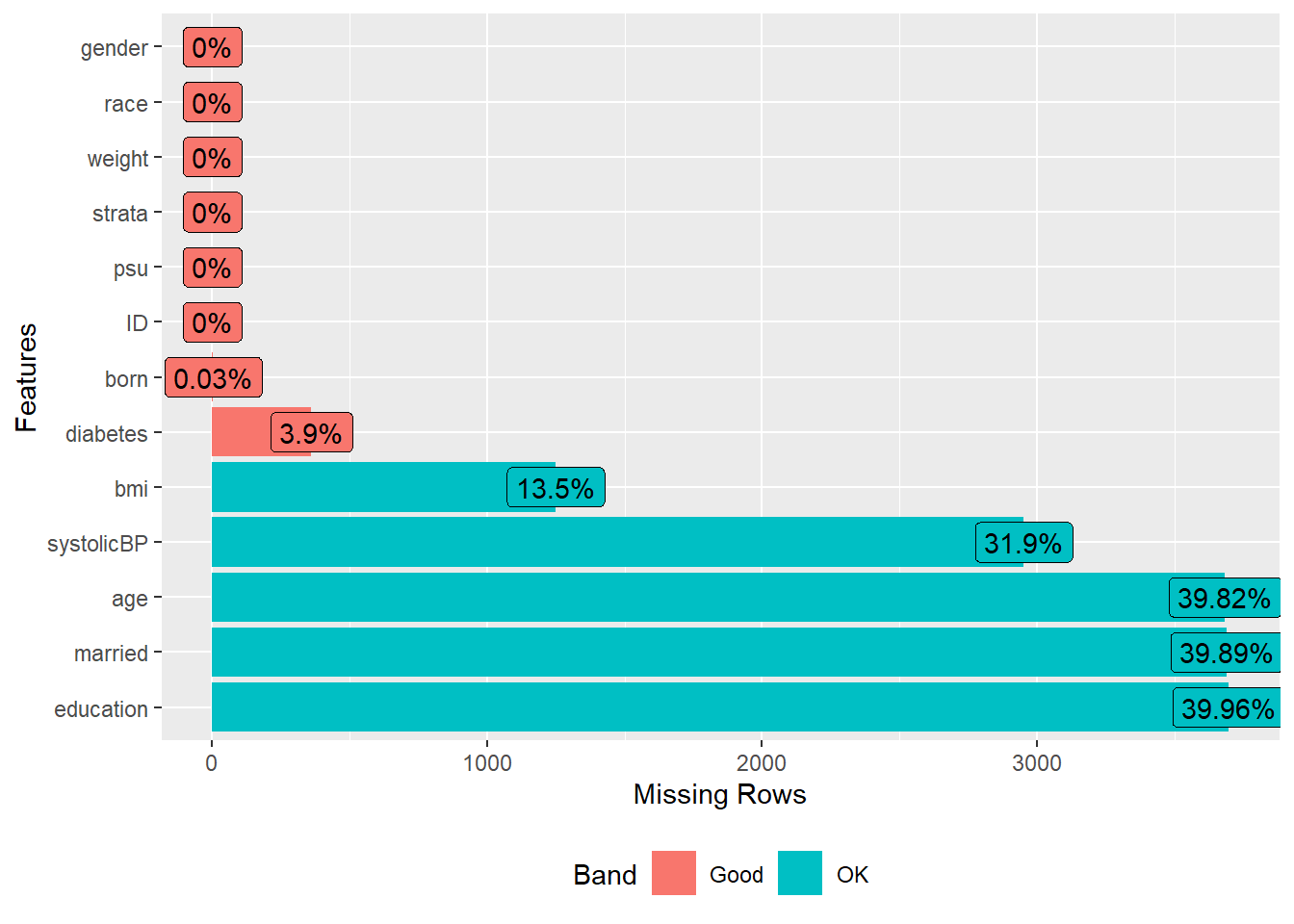
Note that, multiple imputation then delete (MID) approach can be applied if the outcome had some missing values. Due to the small number of missingness, MICE may not impute the outcomes BTW.
Multiple imputation then delete (MID):
MID is a specific approach used in the context of multiple imputation (MI) when dealing with missing outcome data. All missing values, including those in the outcome variable, are imputed to create several complete datasets. In subsequent analyses, the imputed values for the outcome variable are deleted, so that only observed outcome values are analyzed. Each dataset (with observed outcome values and imputed predictor values) is analyzed separately, and results are pooled to provide a single estimate.
Logistic regression
Initialization
The MI process is initialized, setting up the framework for subsequent imputations.
Setting imputation model covariates
The predictor matrix is adjusted to specify which variables will be used to predict missing values in the imputation model. Setting strata as auxiliary variable:
pred <- imputation$pred
pred
#> ID psu strata weight race age married education gender bmi
#> ID 0 1 1 1 1 1 1 1 1 1
#> psu 1 0 1 1 1 1 1 1 1 1
#> strata 1 1 0 1 1 1 1 1 1 1
#> weight 1 1 1 0 1 1 1 1 1 1
#> race 1 1 1 1 0 1 1 1 1 1
#> age 1 1 1 1 1 0 1 1 1 1
#> married 1 1 1 1 1 1 0 1 1 1
#> education 1 1 1 1 1 1 1 0 1 1
#> gender 1 1 1 1 1 1 1 1 0 1
#> bmi 1 1 1 1 1 1 1 1 1 0
#> systolicBP 1 1 1 1 1 1 1 1 1 1
#> born 1 1 1 1 1 1 1 1 1 1
#> diabetes 1 1 1 1 1 1 1 1 1 1
#> systolicBP born diabetes
#> ID 1 1 1
#> psu 1 1 1
#> strata 1 1 1
#> weight 1 1 1
#> race 1 1 1
#> age 1 1 1
#> married 1 1 1
#> education 1 1 1
#> gender 1 1 1
#> bmi 1 1 1
#> systolicBP 0 1 1
#> born 1 0 1
#> diabetes 1 1 0
pred[,"ID"] <- pred["ID",] <- 0
pred[,"psu"] <- pred["psu",] <- 0
pred[,"weight"] <- pred["weight",] <- 0
pred["strata",] <- 0
pred
#> ID psu strata weight race age married education gender bmi
#> ID 0 0 0 0 0 0 0 0 0 0
#> psu 0 0 0 0 0 0 0 0 0 0
#> strata 0 0 0 0 0 0 0 0 0 0
#> weight 0 0 0 0 0 0 0 0 0 0
#> race 0 0 1 0 0 1 1 1 1 1
#> age 0 0 1 0 1 0 1 1 1 1
#> married 0 0 1 0 1 1 0 1 1 1
#> education 0 0 1 0 1 1 1 0 1 1
#> gender 0 0 1 0 1 1 1 1 0 1
#> bmi 0 0 1 0 1 1 1 1 1 0
#> systolicBP 0 0 1 0 1 1 1 1 1 1
#> born 0 0 1 0 1 1 1 1 1 1
#> diabetes 0 0 1 0 1 1 1 1 1 1
#> systolicBP born diabetes
#> ID 0 0 0
#> psu 0 0 0
#> strata 0 0 0
#> weight 0 0 0
#> race 1 1 1
#> age 1 1 1
#> married 1 1 1
#> education 1 1 1
#> gender 1 1 1
#> bmi 1 1 1
#> systolicBP 0 1 1
#> born 1 0 1
#> diabetes 1 1 0Setting imputation model specification
The method for imputing a particular variable is specified (e.g., using Predictive Mean Matching). Here, we add pmm for bmi:
Impute incomplete data
Multiple datasets are imputed, each providing a different “guess” at the missing values, based on observed data. We are imputing m = 3 times.
imputation <- mice(data = dat.with.miss,
seed = 123,
predictorMatrix = pred,
method = meth,
m = 3,
maxit = 5,
print = FALSE)
impdata <- mice::complete(imputation, action="long")
impdata$.id <- NULL
m <- 3
set.seed(123)
allImputations <- imputationList(lapply(1:m,
function(n)
subset(impdata,
subset=.imp==n)))
str(allImputations)
#> List of 2
#> $ imputations:List of 3
#> ..$ :'data.frame': 9254 obs. of 14 variables:
#> .. ..$ ID : int [1:9254] 93703 93704 93705 93706 93707 93708 93709 93710 93711 93712 ...
#> .. ..$ psu : Factor w/ 2 levels "1","2": 2 1 2 2 1 2 1 1 2 2 ...
#> .. ..$ strata : Factor w/ 15 levels "134","135","136",..: 12 10 12 1 5 5 3 1 1 14 ...
#> .. ..$ weight : num [1:9254] 8540 42567 8338 8723 7065 ...
#> .. ..$ race : Factor w/ 4 levels "Black","Hispanic",..: 3 4 1 3 3 3 1 4 3 2 ...
#> .. ..$ age : int [1:9254] 57 46 66 50 23 66 75 49 56 36 ...
#> .. ..$ married : Factor w/ 3 levels "Married","Never.married",..: 1 1 3 1 2 1 3 3 1 1 ...
#> .. ..$ education : Factor w/ 3 levels "College","High.School",..: 1 2 2 1 1 3 1 2 1 3 ...
#> .. ..$ gender : Factor w/ 2 levels "Female","Male": 1 2 1 2 2 1 1 1 2 2 ...
#> .. ..$ bmi : num [1:9254] 17.5 15.7 31.7 21.5 18.1 23.7 38.9 31.2 21.3 19.7 ...
#> .. ..$ systolicBP: int [1:9254] 108 96 200 112 128 124 120 122 108 112 ...
#> .. ..$ born : Factor w/ 2 levels "Born in US","Others": 1 1 1 1 1 2 1 1 2 2 ...
#> .. ..$ diabetes : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 2 1 1 1 1 ...
#> .. ..$ .imp : int [1:9254] 1 1 1 1 1 1 1 1 1 1 ...
#> ..$ :'data.frame': 9254 obs. of 14 variables:
#> .. ..$ ID : int [1:9254] 93703 93704 93705 93706 93707 93708 93709 93710 93711 93712 ...
#> .. ..$ psu : Factor w/ 2 levels "1","2": 2 1 2 2 1 2 1 1 2 2 ...
#> .. ..$ strata : Factor w/ 15 levels "134","135","136",..: 12 10 12 1 5 5 3 1 1 14 ...
#> .. ..$ weight : num [1:9254] 8540 42567 8338 8723 7065 ...
#> .. ..$ race : Factor w/ 4 levels "Black","Hispanic",..: 3 4 1 3 3 3 1 4 3 2 ...
#> .. ..$ age : int [1:9254] 24 49 66 32 34 66 75 80 56 28 ...
#> .. ..$ married : Factor w/ 3 levels "Married","Never.married",..: 2 1 3 2 1 1 3 3 1 2 ...
#> .. ..$ education : Factor w/ 3 levels "College","High.School",..: 1 1 2 1 2 3 1 1 1 1 ...
#> .. ..$ gender : Factor w/ 2 levels "Female","Male": 1 2 1 2 2 1 1 1 2 2 ...
#> .. ..$ bmi : num [1:9254] 17.5 15.7 31.7 21.5 18.1 23.7 38.9 24.9 21.3 19.7 ...
#> .. ..$ systolicBP: int [1:9254] 102 104 136 112 128 120 120 120 108 112 ...
#> .. ..$ born : Factor w/ 2 levels "Born in US","Others": 1 1 1 1 1 2 1 1 2 2 ...
#> .. ..$ diabetes : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 2 1 1 1 1 ...
#> .. ..$ .imp : int [1:9254] 2 2 2 2 2 2 2 2 2 2 ...
#> ..$ :'data.frame': 9254 obs. of 14 variables:
#> .. ..$ ID : int [1:9254] 93703 93704 93705 93706 93707 93708 93709 93710 93711 93712 ...
#> .. ..$ psu : Factor w/ 2 levels "1","2": 2 1 2 2 1 2 1 1 2 2 ...
#> .. ..$ strata : Factor w/ 15 levels "134","135","136",..: 12 10 12 1 5 5 3 1 1 14 ...
#> .. ..$ weight : num [1:9254] 8540 42567 8338 8723 7065 ...
#> .. ..$ race : Factor w/ 4 levels "Black","Hispanic",..: 3 4 1 3 3 3 1 4 3 2 ...
#> .. ..$ age : int [1:9254] 47 71 66 71 45 66 75 37 56 47 ...
#> .. ..$ married : Factor w/ 3 levels "Married","Never.married",..: 1 1 3 1 1 1 3 1 1 1 ...
#> .. ..$ education : Factor w/ 3 levels "College","High.School",..: 1 1 2 1 1 3 1 1 1 2 ...
#> .. ..$ gender : Factor w/ 2 levels "Female","Male": 1 2 1 2 2 1 1 1 2 2 ...
#> .. ..$ bmi : num [1:9254] 17.5 15.7 31.7 21.5 18.1 23.7 38.9 15.9 21.3 19.7 ...
#> .. ..$ systolicBP: int [1:9254] 100 114 162 112 128 166 120 116 108 112 ...
#> .. ..$ born : Factor w/ 2 levels "Born in US","Others": 1 1 1 1 1 2 1 1 2 2 ...
#> .. ..$ diabetes : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 2 1 1 1 1 ...
#> .. ..$ .imp : int [1:9254] 3 3 3 3 3 3 3 3 3 3 ...
#> $ call : language imputationList(lapply(1:m, function(n) subset(impdata, subset = .imp == n)))
#> - attr(*, "class")= chr "imputationList"Design
A survey design object is created, ensuring that subsequent analyses appropriately account for the survey design.
Survey data analysis
A logistic regression model is fitted to each imputed dataset.
model.formula <- as.formula(I(diabetes == 'Yes') ~
born + race + age + married +
education + gender + bmi + systolicBP)
fit.from.logistic <- with(w.design, svyglm(model.formula, family = binomial("logit")))
#> Warning in eval(family$initialize): non-integer #successes in a binomial glm!
#> Warning in eval(family$initialize): non-integer #successes in a binomial glm!
#> Warning in eval(family$initialize): non-integer #successes in a binomial glm!Pooled estimates
Results from models across all imputed datasets are pooled to provide a single estimate, accounting for the uncertainty due to missing data.
pooled.estimates <- MIcombine(fit.from.logistic)
summary(pooled.estimates, digits = 2, logeffect=TRUE)
#> Multiple imputation results:
#> with(w.design, svyglm(model.formula, family = binomial("logit")))
#> MIcombine.default(fit.from.logistic)
#> results se (lower upper) missInfo
#> (Intercept) 0.00013 7.5e-05 3.9e-05 0.00041 22 %
#> bornOthers 1.44729 2.7e-01 1.0e+00 2.07384 0 %
#> raceHispanic 0.81619 1.1e-01 6.3e-01 1.05882 0 %
#> raceOther 1.43817 2.6e-01 1.0e+00 2.04954 3 %
#> raceWhite 0.86411 1.3e-01 6.5e-01 1.14994 3 %
#> age 1.06157 3.6e-03 1.1e+00 1.06874 6 %
#> marriedNever.married 0.83242 1.6e-01 5.7e-01 1.20809 10 %
#> marriedPreviously.married 0.88401 1.1e-01 6.8e-01 1.14163 11 %
#> educationHigh.School 1.16331 1.9e-01 8.4e-01 1.60803 0 %
#> educationSchool 1.41943 2.4e-01 1.0e+00 1.98397 7 %
#> genderMale 1.53458 1.8e-01 1.2e+00 1.94217 3 %
#> bmi 1.10597 1.2e-02 1.1e+00 1.12956 1 %
#> systolicBP 1.00325 3.2e-03 1.0e+00 1.01001 39 %
OR <- round(exp(pooled.estimates$coefficients), 2)
OR <- as.data.frame(OR)
CI <- round(exp(confint(pooled.estimates)), 2)
OR <- cbind(OR, CI)
OR[2,]Propensity score matching analysis
Initialization
The MI process is re-initialized to facilitate PSM in the context of MI.
Zanutto E. L. (2006) under multiple imputation
An iterative process is performed within each imputed dataset, which involves:
- Estimating propensity scores.
- Matching treated and untreated subjects based on these scores.
- Extracting matched data and checking the balance of covariates across matched groups.
- Fitting outcome models to the survey weighted matched data and estimating treatment effects.
Notice that we are performing multi-step process within MI
match.statm <- SMDm <- tab1m <- vector("list", m)
fit.from.PS <- vector("list", m)
for (i in 1:m) {
analytic.i <- allImputations$imputations[[i]]
# Rename the weight variable into survey.weight
names(analytic.i)[names(analytic.i) == "weight"] <- "survey.weight"
# Specify the PS model to estimate propensity scores
ps.formula <- as.formula(I(born=="Others") ~
race + age + married + education +
gender + bmi + systolicBP)
# Propensity scores
ps.fit <- glm(ps.formula, data = analytic.i, family = binomial("logit"))
analytic.i$PS <- fitted(ps.fit)
# Match exposed and unexposed subjects
set.seed(123)
match.obj <- matchit(ps.formula, data = analytic.i,
distance = analytic.i$PS,
method = "nearest",
replace = FALSE,
caliper = 0.2,
ratio = 1)
match.statm[[i]] <- match.obj
analytic.i$PS <- match.obj$distance
# Extract matched data
matched.data <- match.data(match.obj)
# Balance checking
cov <- c("race", "age", "married", "education", "gender", "bmi", "systolicBP")
tab1m[[i]] <- CreateTableOne(strata = "born",
vars = cov, data = matched.data,
test = FALSE, smd = TRUE)
SMDm[[i]] <- ExtractSmd(tab1m[[i]])
# Setup the design with survey features
analytic.i$matched <- 0
analytic.i$matched[analytic.i$ID %in% matched.data$ID] <- 1
# Survey setup for full data
w.design0 <- svydesign(strata = ~strata, id = ~psu, weights = ~survey.weight,
data = analytic.i, nest = TRUE)
# Subset matched data
w.design.m <- subset(w.design0, matched == 1)
# Outcome model (double adjustment)
out.formula <- as.formula(I(diabetes == "Yes") ~
born + race + age + married +
education + gender + bmi + systolicBP)
fit.from.PS[[i]] <- svyglm(out.formula, design = w.design.m,
family = quasibinomial("logit"))
}Check matched data
The matched data is inspected to ensure that matching was successful and appropriate.
match.statm
#> [[1]]
#> A `matchit` object
#> - method: 1:1 nearest neighbor matching without replacement
#> - distance: User-defined [caliper]
#> - caliper: <distance> (0.044)
#> - number of obs.: 9254 (original), 3590 (matched)
#> - target estimand: ATT
#> - covariates: race, age, married, education, gender, bmi, systolicBP
#>
#> [[2]]
#> A `matchit` object
#> - method: 1:1 nearest neighbor matching without replacement
#> - distance: User-defined [caliper]
#> - caliper: <distance> (0.044)
#> - number of obs.: 9254 (original), 3598 (matched)
#> - target estimand: ATT
#> - covariates: race, age, married, education, gender, bmi, systolicBP
#>
#> [[3]]
#> A `matchit` object
#> - method: 1:1 nearest neighbor matching without replacement
#> - distance: User-defined [caliper]
#> - caliper: <distance> (0.044)
#> - number of obs.: 9254 (original), 3594 (matched)
#> - target estimand: ATT
#> - covariates: race, age, married, education, gender, bmi, systolicBPCheck balance in matched data
The balance of covariates across matched groups is assessed to ensure that matching has successfully reduced bias.
SMDm
#> [[1]]
#> 1 vs 2
#> race 0.028883793
#> age 0.033614763
#> married 0.007318561
#> education 0.117536503
#> gender 0.040145831
#> bmi 0.043350560
#> systolicBP 0.054549772
#>
#> [[2]]
#> 1 vs 2
#> race 0.019901420
#> age 0.016267050
#> married 0.017196043
#> education 0.128811588
#> gender 0.003338016
#> bmi 0.057014434
#> systolicBP 0.071553721
#>
#> [[3]]
#> 1 vs 2
#> race 0.04490482
#> age 0.01959377
#> married 0.03687394
#> education 0.13301810
#> gender 0.01225625
#> bmi 0.03697878
#> systolicBP 0.10025529Pooled estimate
Finally, the treatment effect estimates from the matched analyses across all imputed datasets are pooled to provide a single, overall estimate, ensuring that the final result appropriately accounts for the uncertainty due to both the matching process and the imputation of missing data.
pooled.estimates <- MIcombine(fit.from.PS)
summary(pooled.estimates, digits = 2, logeffect=TRUE)
#> Multiple imputation results:
#> MIcombine.default(fit.from.PS)
#> results se (lower upper) missInfo
#> (Intercept) 8.9e-05 4.9e-05 0.00003 0.00026 8 %
#> bornOthers 2.0e+00 3.1e-01 1.47719 2.73325 16 %
#> raceHispanic 7.0e-01 1.7e-01 0.42593 1.15504 27 %
#> raceOther 1.4e+00 4.1e-01 0.77209 2.53278 26 %
#> raceWhite 4.9e-01 2.7e-01 0.15853 1.52308 28 %
#> age 1.1e+00 4.6e-03 1.04472 1.06298 8 %
#> marriedNever.married 5.9e-01 2.0e-01 0.28824 1.18926 38 %
#> marriedPreviously.married 1.0e+00 2.5e-01 0.62417 1.64438 11 %
#> educationHigh.School 1.4e+00 3.0e-01 0.89403 2.10852 2 %
#> educationSchool 1.3e+00 3.4e-01 0.81042 2.21438 7 %
#> genderMale 1.3e+00 2.3e-01 0.86695 1.83880 31 %
#> bmi 1.1e+00 1.1e-02 1.08062 1.12285 5 %
#> systolicBP 1.0e+00 3.0e-03 1.00314 1.01494 3 %
OR <- round(exp(pooled.estimates$coefficients), 2)
OR <- as.data.frame(OR)
CI <- round(exp(confint(pooled.estimates)), 2)
OR <- cbind(OR, CI)
OR[2,]