load("data/analytic3cycles.RData")4 Step 1: Proxy sources
We saved analytic3cycles.RData in Appendix. This contained the following datasets:
data.merged= merged data from 3 cycles,data.complete= complete case merged data,dat.proxy.long= only proxy information
4.1 Data with investigator-specified variables
We will work with the data.complete data for the investigator-specified information.
analytic <- data.complete
idx <- analytic$id
outcome <- as.numeric(analytic$diabetes == "Yes")
exposure <- as.numeric(analytic$obese == "Yes")
domain <- "dx"
analytic.dfx <- as.data.frame(cbind(idx, exposure, outcome, domain))We prepare the minimal analytic data only with the following 4 information:
- identifying information (
idx) - exposure (
obese) - outcome (
diabetes) - domain of the codes (
dx). In this example we only have prescription domain (1 domaindx)
4.2 Proxy data
4.2.1 Identify the data dimensions (proxy sources)
In this example we only have prescription domain (1 domain dx of ICD-10-CM code). Hence \(p = 1\) in this exercise.
NHANES Questionnaire collects information on: (a) dietary supplements, (b) nonprescription antacids, (c) prescription medications, and (d) preventive aspirin use.
4.2.2 Define a covariate assessment period (CAP)

We only collect proxy information from a well-defined CAP. In our case, it was \(30\) days.
NHANES asked “In the past 30 days, have you used or taken medication for which a prescription is needed? Do not include prescription vitamins or minerals you may have already told me about.”
We will work with the merge proxy data (ICD-10 codes) from 3 cycles: dat.proxy.long.
4.2.3 Omit duplicated information
We need to delete codes that could be close proxies of exposure and/or outcome, or other investigator specified covariates we have already selected in step0.
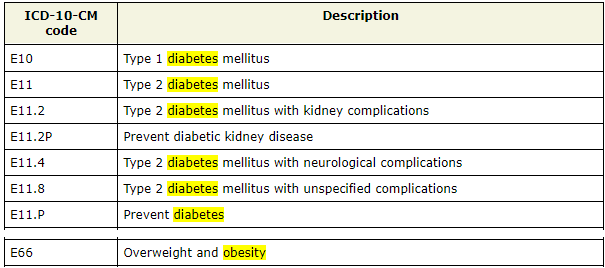
dat.proxy.long <- subset(dat.proxy.long,
icd10 != "E66") # Overweight and obesity
dat.proxy.long <- subset(dat.proxy.long,
icd10 != "O24") # Gestational diabetes mellitus
dat.proxy.long <- subset(dat.proxy.long,
icd10 != "E10") # Type 1 diabetes mellitus
dat.proxy.long <- subset(dat.proxy.long,
icd10 != "E11") # Type 2 diabetes mellitus- We delete codes associated with exposure and outcome.
- Same should be done for any other proxies that may have duplicating information compared to the investigator-specified covariates.
4.2.4 Long format proxy data
Here is an example of 3 digit codes for 1 patient with subject ID “100001”. We create the same for all patients.
| ID | ICD 10 codes (3 digit) | Description |
|---|---|---|
| 100001 | F33 | Major depressive disorder, recurrent |
| 100001 | I10 | Hypertension |
| 100001 | M62 | Muscle spasm |
| 100001 | F32 | Major depressive disorder, single episode |
| 100001 | M25 | Joint disorder/pain |
| 100001 | K21 | Gastro-esophageal reflux disease |
| 100001 | M79 | musculoskeletal pain conditions |
| 100001 | R12 | Heartburn |
4.3 Merge Proxy data with Analytic data
- We will work with the merge proxy data with analytic data.
- That will provide us with the IDs (
idx) of the subject that have proxy (ICD-10) information associated with them.
require(dplyr)
dfx <- merge(analytic.dfx, proxy.var.long, by = "idx")
head(dfx)basetable <- dfx %>% select(idx, exposure, outcome) %>% distinct()
patientIds <- basetable$idx
length(patientIds)
#> [1] 7585