flowchart LR S(Super Learner) --> l(Logistic regression) S --> g(LASSO) S --> m(Multivariate Adaptive Regression Splines MARS) style S fill:#90EE90;
We show an example using a super learner using 3 candidate learners.
If you want to know more about Super Learner, look at other tutorials.
flowchart LR S(Super Learner) --> l(Logistic regression) S --> g(LASSO) S --> m(Multivariate Adaptive Regression Splines MARS) style S fill:#90EE90;
The super learning approach is fundamentally different from the pure ML or LASSO approach discussed earlier. Here all of the candidate learners are using exposure as their outcome while running the model.
proxy.list <- names(out2[-1])
# out3$autoselected_covariate_df[,-1] for hybrid
# out2 is from step2$recurrence_data
length(proxy.list)
#> [1] 91
covform <- paste0(investigator.specified.covariates, collapse = "+")
proxyform <- paste0(proxy.list, collapse = "+")
rhsformula <- paste0(c(covform, proxyform), collapse = "+")
ps.formula <- as.formula(paste0("exposure", "~", rhsformula))We work with all proxies
require(WeightIt)
W.out <- weightit(ps.formula,
data = hdps.data,
estimand = "ATE",
method = "super",
SL.library = c("SL.glm",
"SL.glmnet",
"SL.earth"))
#> Loading required namespace: glmnet
#> Loading required namespace: earth
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurredPropensity score model fit based on super learning algorithm to be able to calculate the inverse probability weights.
summary(W.out$ps)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.01919 0.22474 0.38968 0.42094 0.59528 0.99131
out.formula <- as.formula(paste0("outcome", "~", "exposure"))
fit <- glm(out.formula,
data = hdps.data,
weights = W.out$weights,
family= binomial(link = "logit"))
fit.summary <- summary(fit)$coef["exposure",
c("Estimate",
"Std. Error",
"Pr(>|z|)")]
fit.summary[2] <- sqrt(sandwich::sandwich(fit)[2,2])
require(lmtest)
conf.int <- confint(fit, "exposure", level = 0.95, method = "hc1")
fit.summary_with_ci.sl <- c(fit.summary, conf.int)
knitr::kable(t(round(fit.summary_with_ci.sl,2))) | Estimate | Std. Error | Pr(>|z|) | 2.5 % | 97.5 % |
|---|---|---|---|---|
| 0.43 | 0.09 | 0 | 0.32 | 0.54 |
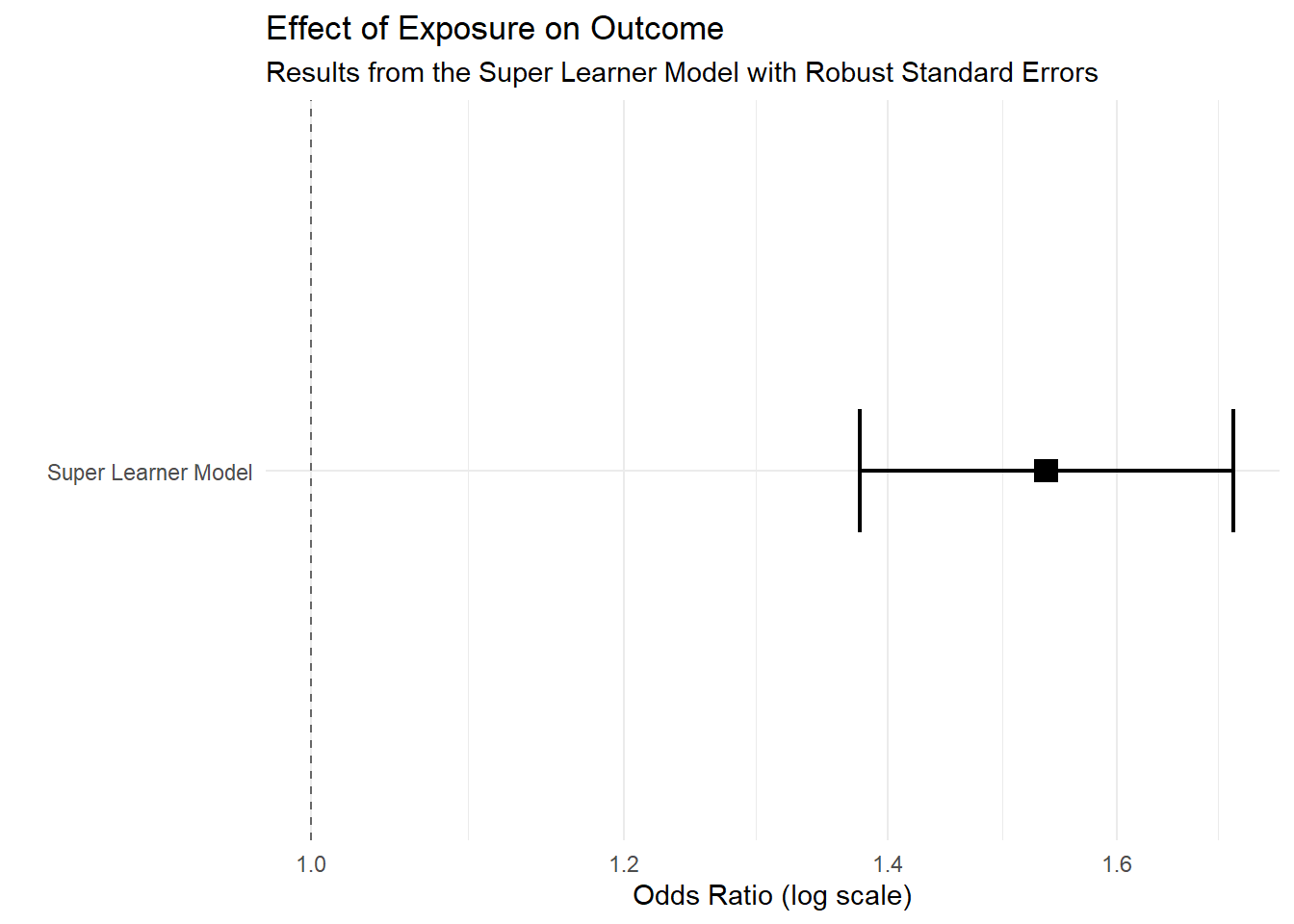
Summary of results (log-OR).